GRANAR

GRANAR, the Generator of Root ANAtomy in R, is a computational plant cell scale model designed to simulate root anatomical networks. These networks are crafted using easily accessible anatomical features, which can be obtained from open-access image analysis software and root cross-section images. Once the root anatomy is generated, it can be saved as an XML file. The model can simulate different vascular patterns, depending on whether it’s modeling a monocot or dicot root. You can think of GRANAR like a computer program that can recreate the structure of plant roots on a cell-by-cell basis. Explore more about GRANAR here.
MECHA
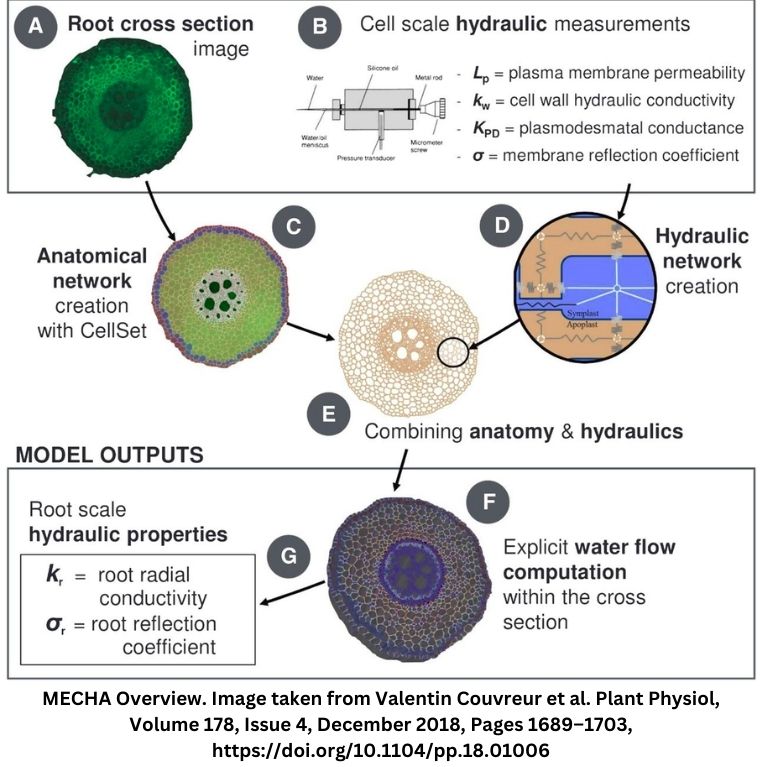
MECHA is a mathematical plant organ scale model designed to compute the flow of water through various parts of individual cells in a complete root cross-section. This includes the cell walls, membranes, and plasmodesmata. The model takes into account detailed root anatomical descriptions and a minimal set of cell-level hydraulic properties. It uses experimental data on the permeability of cell walls, membranes, and plasmodesmata. The model also considers hydraulic principles at both the cell and root segment scales. Explore more about MECHA here.
CPlantBox
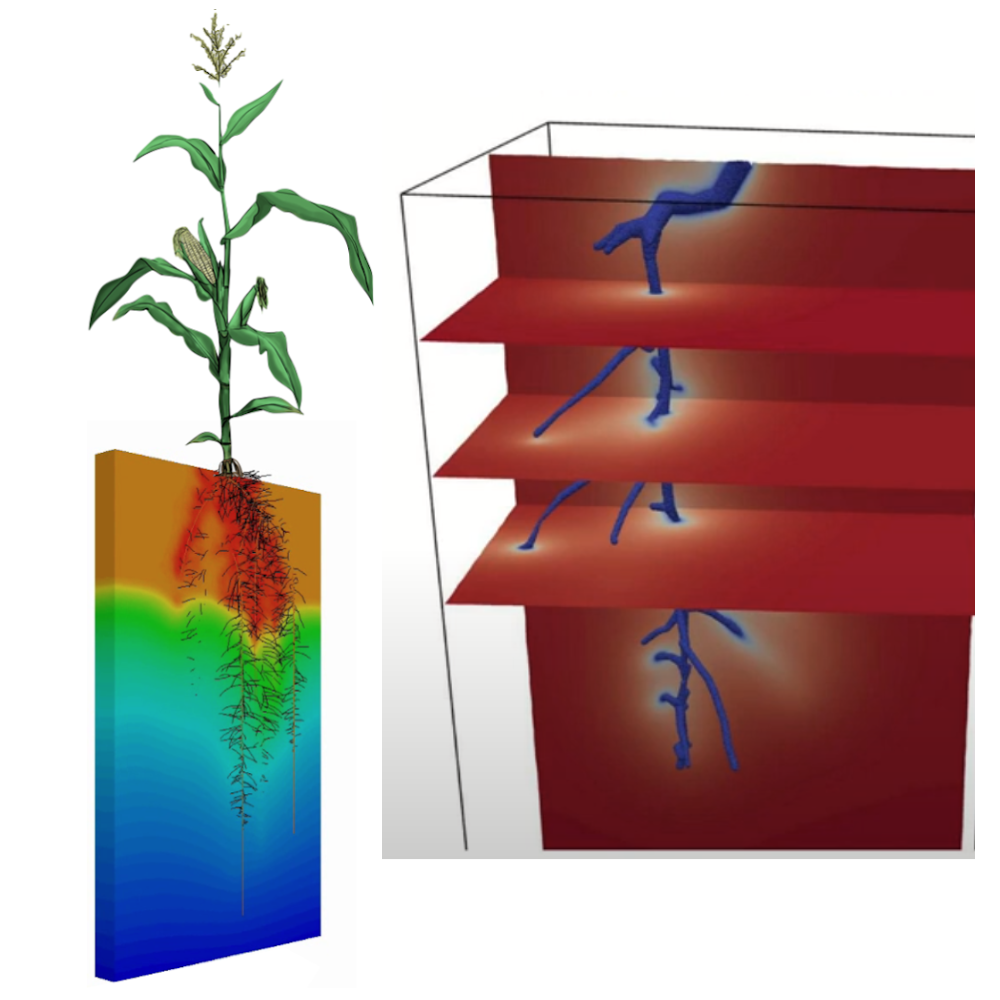
CPlantBox is a full plant modeling framework that focuses on the growth and development of plant architectures, including both the root and shoot systems. It represents the plant as a single topological network of organs. The simulated plant architecture is made up of nodes or coordinates, and these nodes’ properties and interactions form the entire network. For the root part, CPlantBox inherits the flexibility of CRootBox, allowing it to generate any type of root architecture. For the shoot, it has implemented various branching and leaf arrangement patterns. By combining these patterns, CPlantBox can simulate many types of shoot architectures. Further, the latest CPlantBox implementation provides linkage with different modules (photosynthesis and carbon flow) within a single framework, enhancing its capabilities and making it a versatile tool for plant studies. Explore more about CPlantBox here.
AgroC
AgroC is a crop-scale 1D soil-column model designed for simulating crucial aspects of crop growth and soil interactions, including nutrient dynamics, water fluxes, and carbon cycling. The model is tailored for in-depth understanding of crop and soil behavior under various environmental conditions. It accurately models soil carbon turnover, CO2 flux, and plant water stress. Unique features like root exudation and death are considered, highlighting its advanced capabilities in representing real-world agricultural scenarios.Explore more about AgroC here.
SIMPLACE
SIMPLACE is a crop-scale model that helps understand complex interactions between crops, soil, and climate. By coupling different modules, each representing a process like water use or plant growth, SIMPLACE offers customizable solutions for different agricultural needs.Its multi-threaded high-performance architecture allows for calibration and simulations at varying spatial scales.Explore more about SIMPLACE here.

TerrSysMP
TerrSysMP (Terrestrial Systems Modeling Platform) is a robust, open-source regional scale framework designed for simulating complex interactions between the atmosphere, land surface, and subsurface. It integrates the COSMO or ICON atmospheric models, the Community Land Model (CLM), and the ParFlow model for a holistic view of environmental dynamics, particularly in the hydrological cycle.Explore more about TerrSysMP here